Chapter 3 Getting started
3.1 Requirements
FSL for FLIRT
Oryx-MRSI is tested on MAC (2.9 GHz Quad-Core Intel Core i7, 16 GB 2133 MHz LPDDR3, Radeon Pro 560 4 GB Intel HD Graphics 630 1536 MB ) and Ubuntu 18.04.4 LTS (Memory 32GIB, Processor Intel Core i7-9800X CPU (3.8GHzx16?), Graphics GeForce RTX 2070/PCle/SSE2)
3.2 Install Oryx-MRSI
Oryx-MRSI uses FSL-Flirt function so using FSL from MATLAB should be ready.
If you want to install FSL into your computer, check this link
If you use MAC, check this link (Advance Usage part-Using FSL from MATLAB)
If you use LINUX, check this link (Using FSL from MATLAB)
Plase download SPM12 using this link
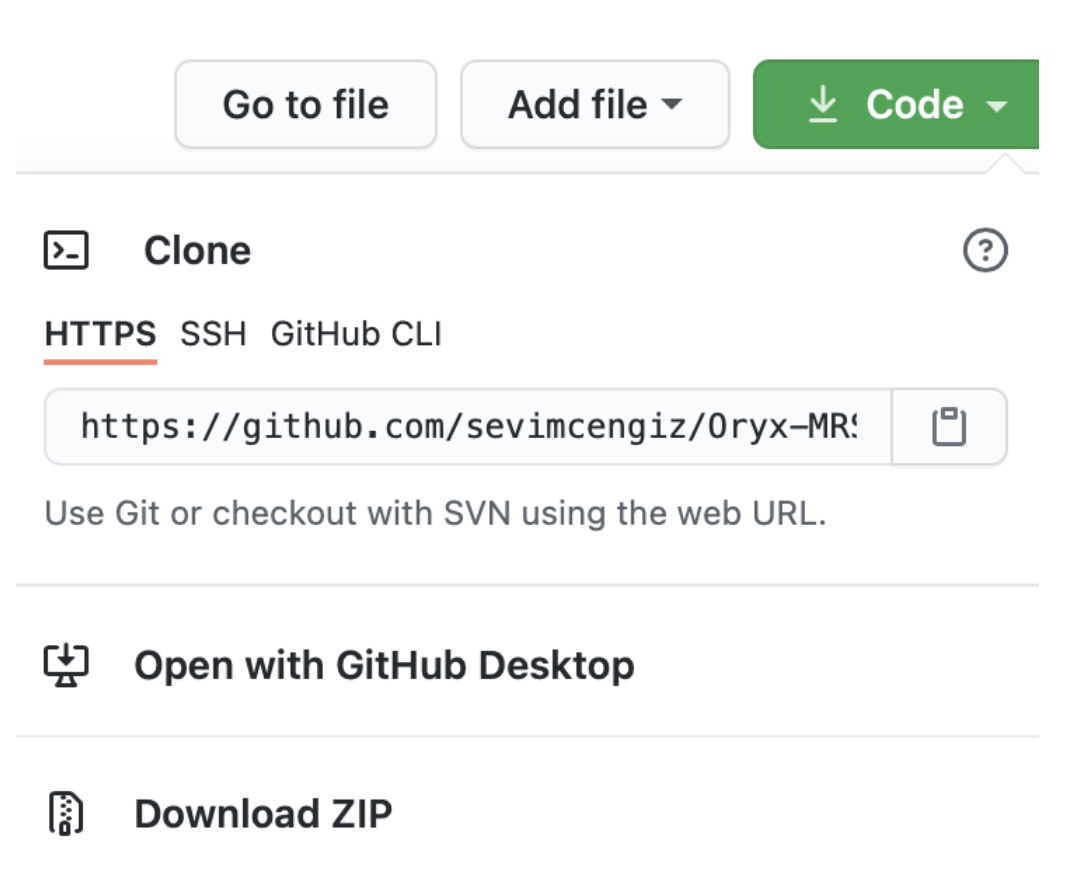
Please, check Oryx-MRSI Github Repository.
Please, clone the repository, or download. See the figure below.
3.3 Before Use Oryx-MRSI
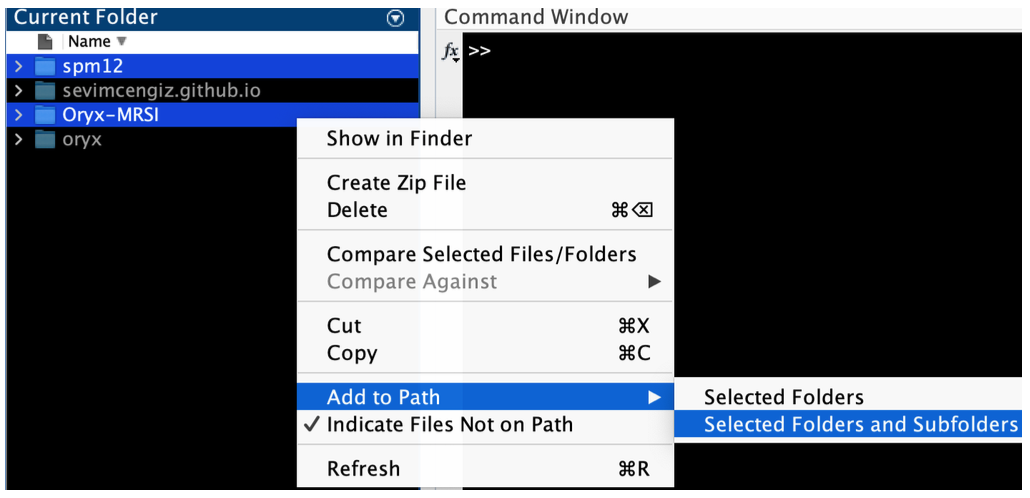
Addpath Oryx-MRSI with subfolders.
Addpath SPM12 with subfolders.
Please make sure that FSL usage from Matlab command window installation is completed properly.
Before running a data analysis using Oryx-MRSI, let’s check that FSL usage is from Matlab is done.
Please open matlab and run check_fsl_usage_from_matlab.m script which is given under Oryx-MRSI Github repo.
If there is no error, FSL usage from Matlab is completely installed.
If you get an error, plese check these:
If you use MAC, check this link (Advance Usage part-Using FSL from MATLAB)
If you use LINUX, check this link (Using FSL from MATLAB)
3.4 How to organize your raw data
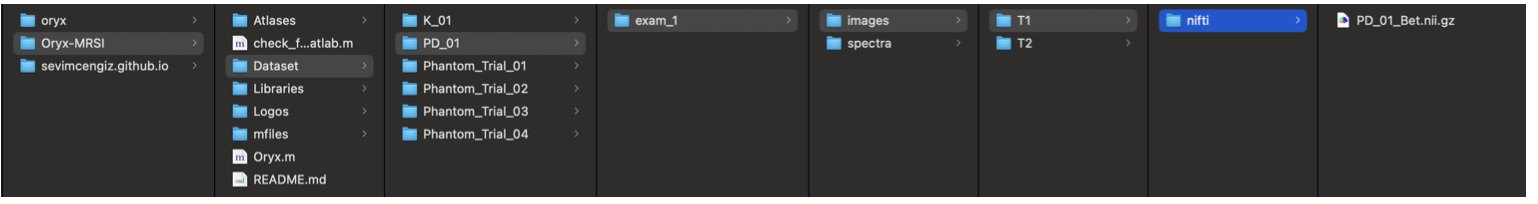
If you want to go through with your own data set, the folder of data structure given below will help you.
There is a dataset folder under Oryx-MRSI.
Each patient should be listed under that folder.
Each patient should have T1w-MRI, T2w-MRI or both.
If you have T1w-MRI,
- ~/Oryx-MRSI/Dataset/PatientName/exam_1/images/T1/nifti/ PatientName_Bet.nii.gz
If you have T2w-MRI,
- ~/Oryx-MRSI/Dataset/PatientName/exam_1/images/T2/nifti/ PatientName_Bet.nii.gz
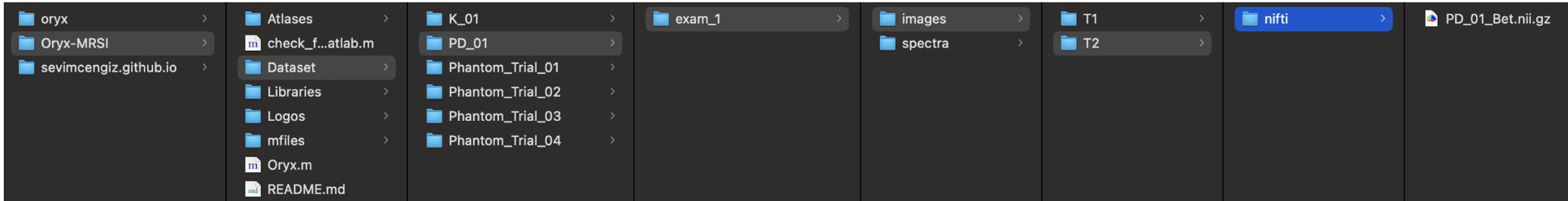
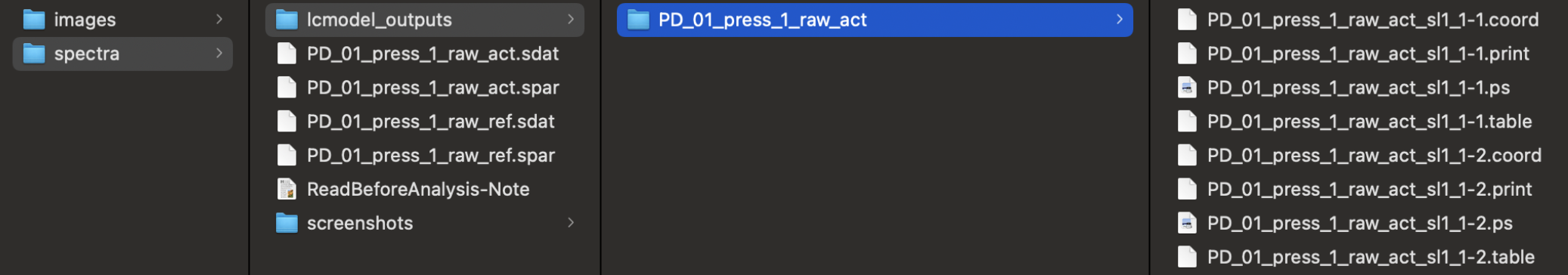
You need Spar/MRS-NIfTI data and LCModel outputs.
For spectra, - ~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/example-MRS.nii
~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/PatientName_press_1_raw_act.sdat
~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/PatientName_press_1_raw_act.spar
~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/PatientName_press_1_raw_ref.sdat
~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/PatientName_press_1_raw_ref.spar
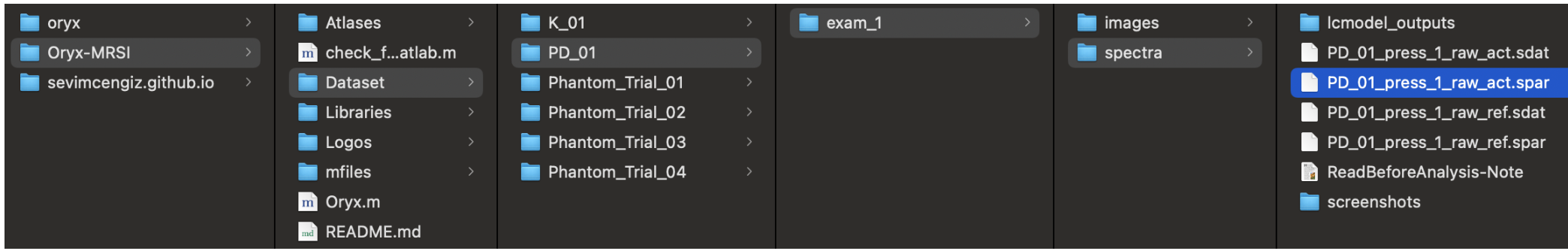
For LCModel outputs at press_1;
- ~/Oryx-MRSI/Dataset/PatientName/exam_1/spectra/lcmodel_outputs/PatientName_press_1_raw_act/
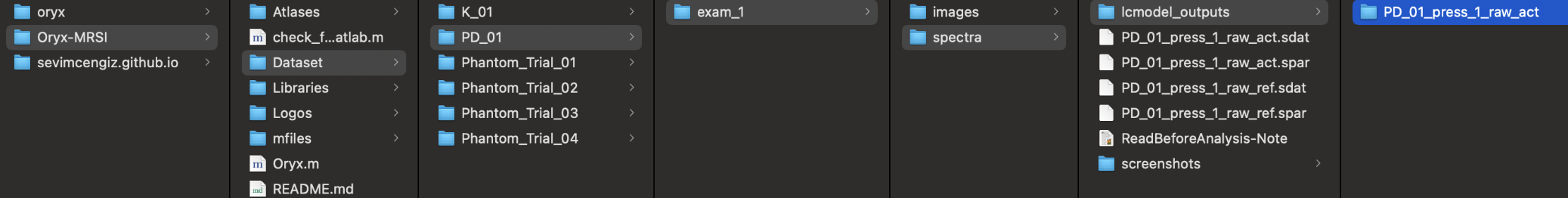
Each folder has to have .coord and .table files. They need to be named as;
PatientName_press_1_raw_act_sl1_4-7.coord (It shows slice 1, row 4, column 7)
PatientName_press_1_raw_act_sl1_4-7.table (It shows slice 1, row 4, column 7)